XU Baofu, Ph.D. Professor
Baofu Xu obtained his B.S. in bioengineering from Shandong Agricultural University. He received his Ph.D. from CAS Center for Excellence in Molecular Plant Sciences/Institute of Plant Physiology and Ecology where he worked to elucidate the biosynthetic pathways of plant-derived natural products with multi-omics and biochemical techniques under the supervision of Prof. Youli Xiao . He then started working as a postdoctoral associate with Prof. Jeffrey D. Rudolf at the University of Florida, focusing on bacterial natural product discovery, pathway elucidation, and enzymology. Currently, he is working as a Prof. at the Shanghai Institute of Materia Medica (Yantai Advanced Research Institute), Chinese Academy of Sciences. The main research interests of his lab are natural product chemistry, genome mining, and biosynthesis-related studies. In his spare time, he enjoys biking, running, and meditation.
Research Areas
Research Interests
A combination of informatics, biology, and chemistry drives the discovery of new medicines
1. DISCOVER: Genome mining-based discovery of lead compounds
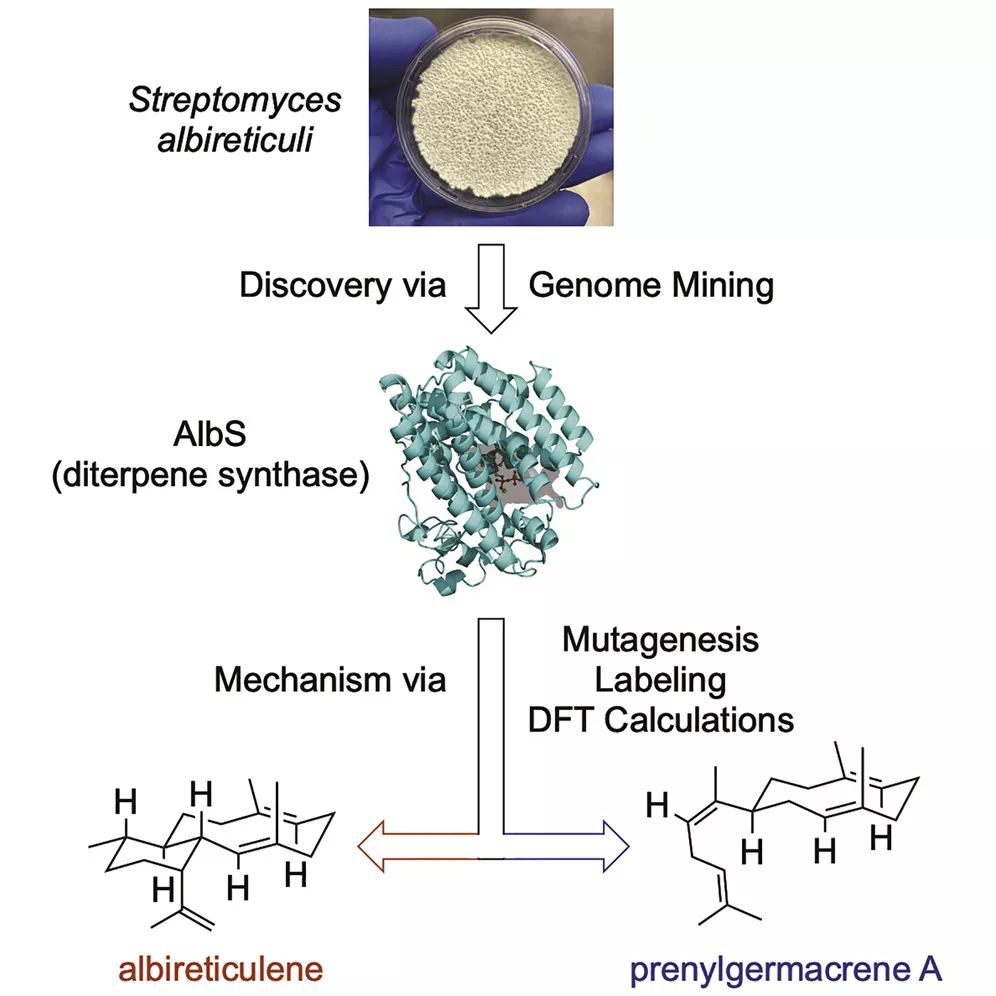
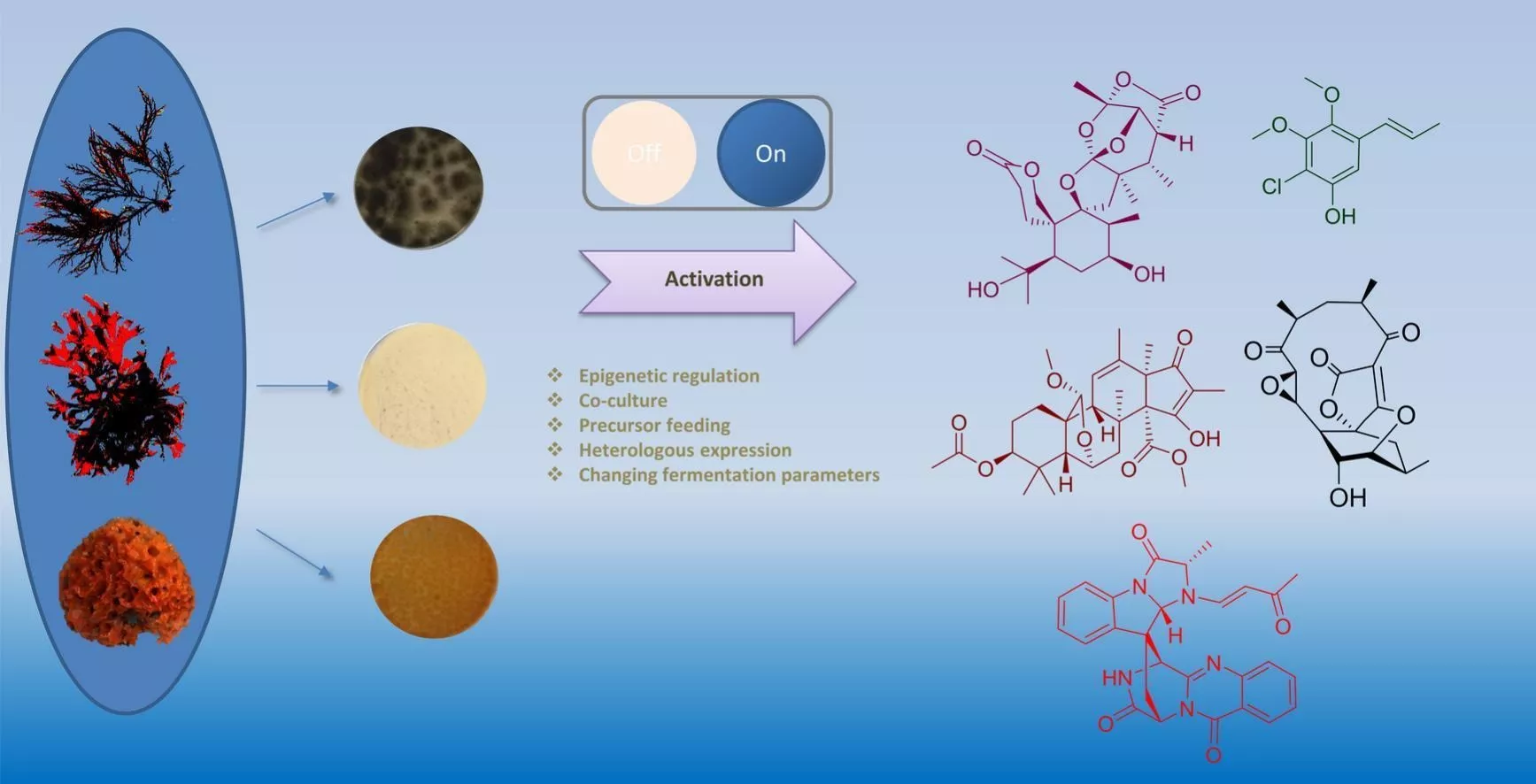
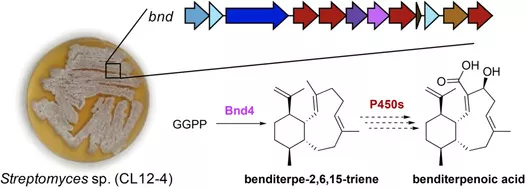
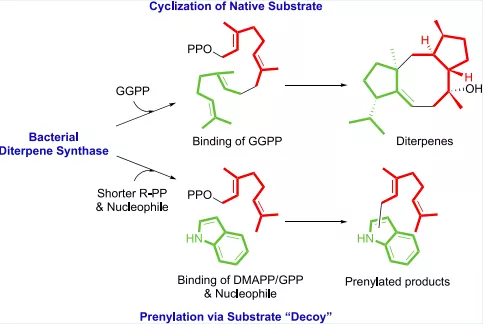
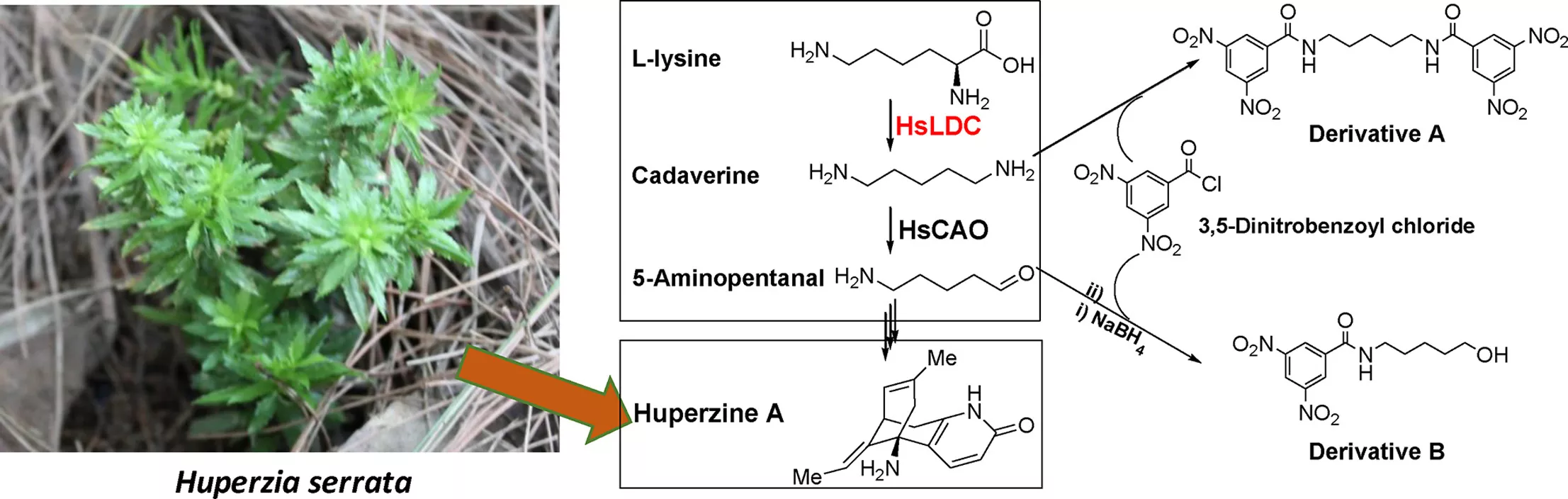
Natural products are the main sources of pharmaceuticals. The nature of natural productsis a series of enzymes encoded in the genome of living material that are doing various chemical reactions, leading to the synthesis of a large number of defensive compounds.In other words, genomes contain the mystery of natural product biosynthesis. In return, the genomes could facilitate the discovery of new compounds in terms of new core scaffolds or functionalization (types or positions). Currently, the computer-based discovery of unexplored Biosynthesis Gene Clusters (BGC) is rather simple in terms of genome mining with “core genes”, “resistant genes”, “modifying genes”, etc. with the help of user-friendly web tools, e.g., antiSMASH, EFI-EST, Operon Mapper, and the help of ever-growing genome database. And large quantities of hidden BGCs can be discovered and wait to be explored. However, activation of the BGC to synthesize new compounds is much more difficult than the virtual mining process. Thus, the primary task of our lab is to develop a set of genetic toolboxes for activating BGCs to high-efficiently turn virtual mining into real compounds.
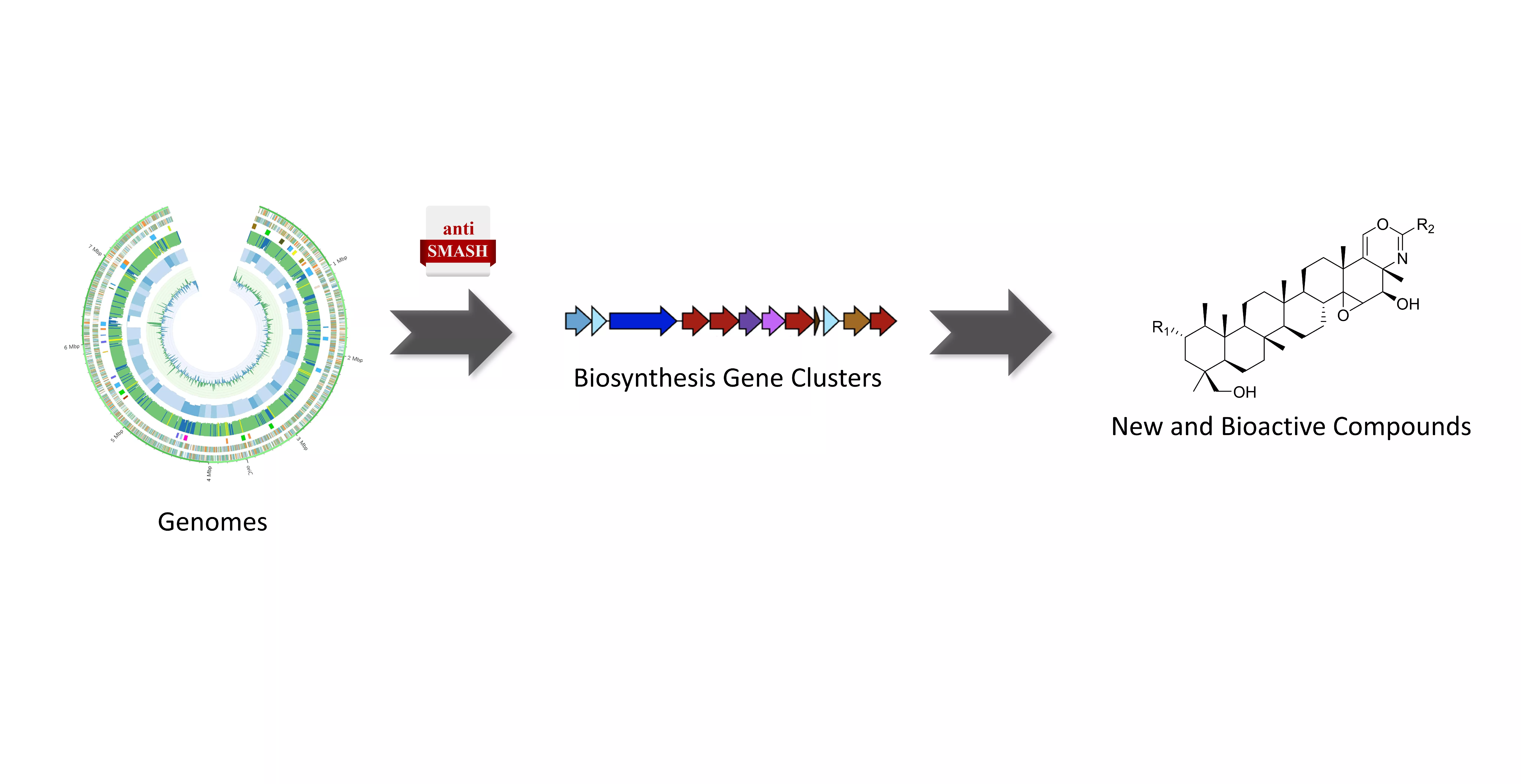
2. ELUCIDATE: Biosynthetic pathway elucidation of bioactive compounds
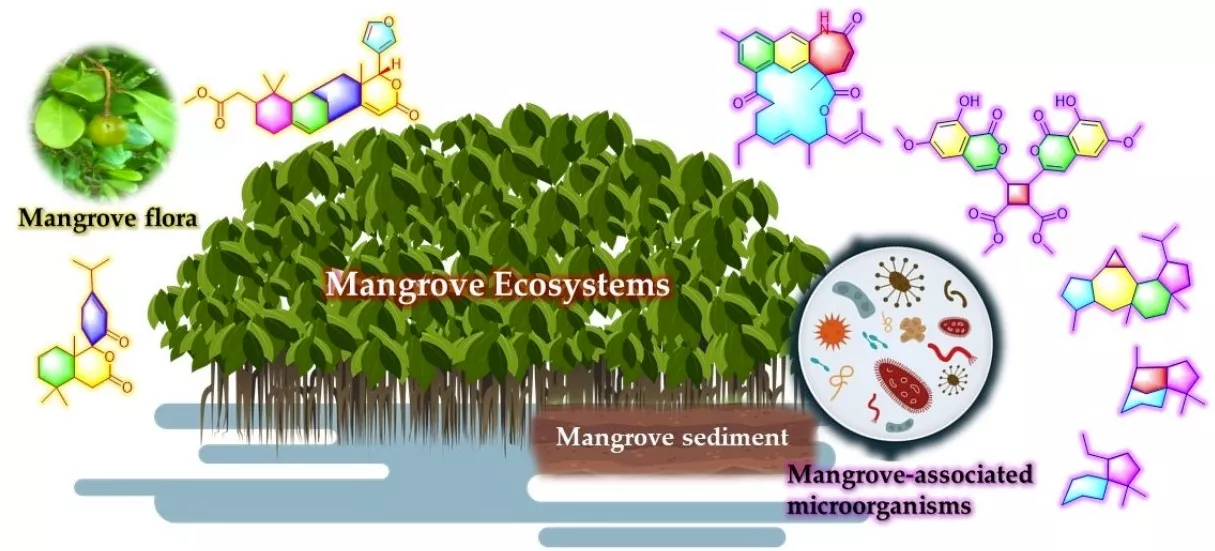
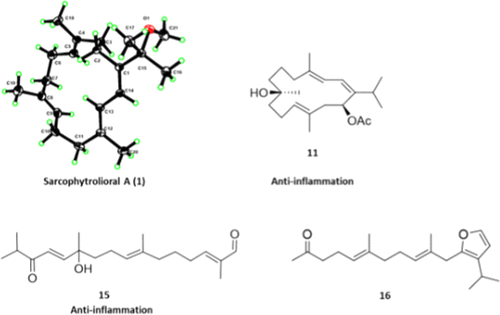
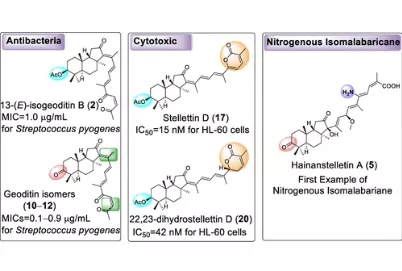
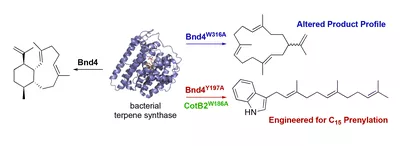
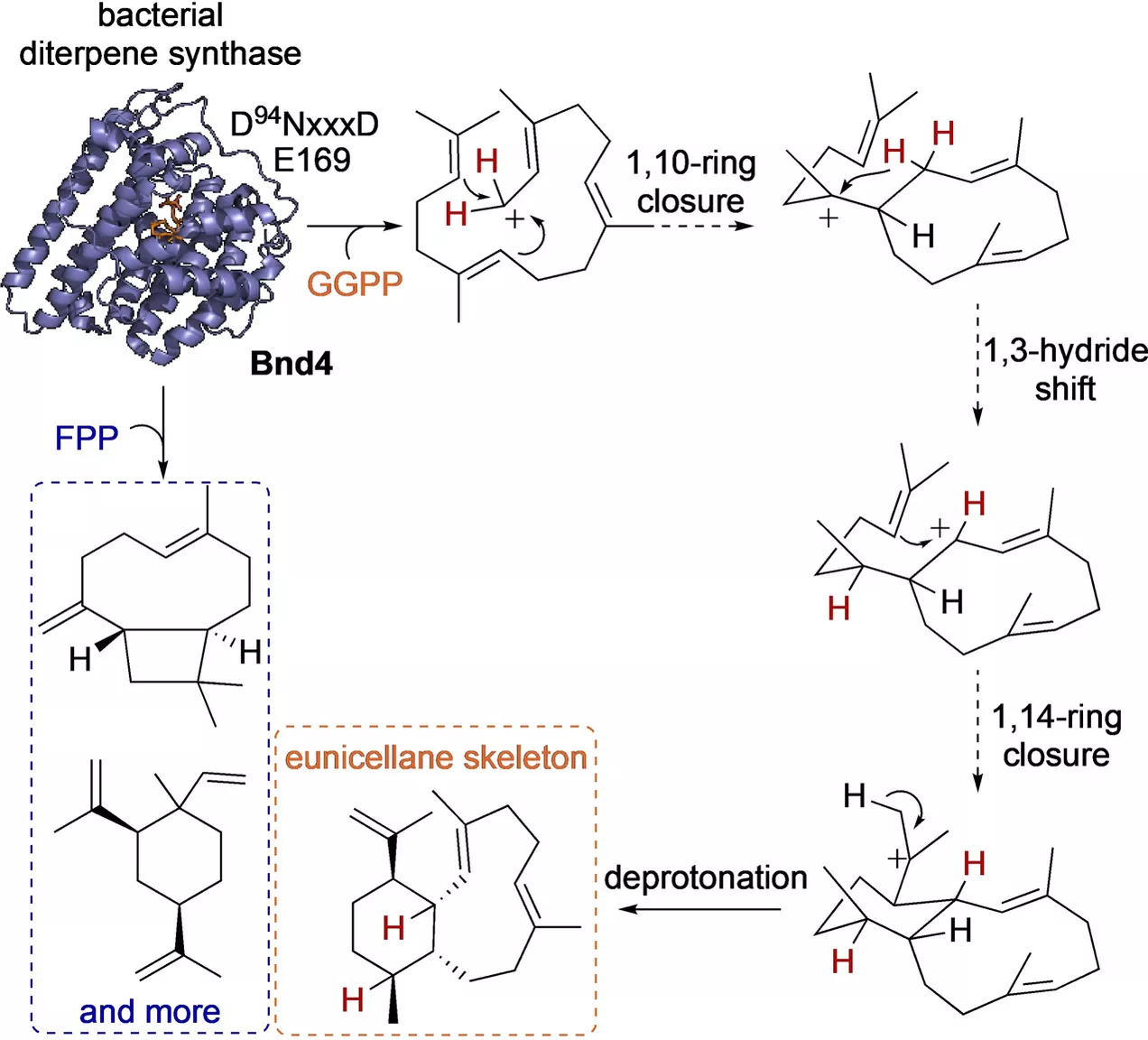
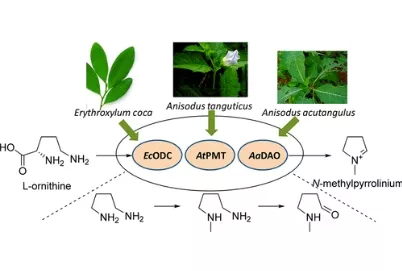
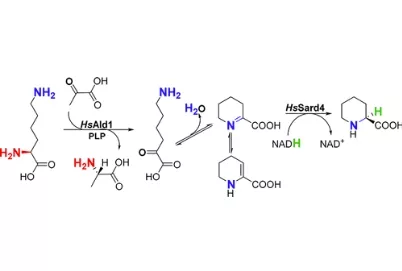
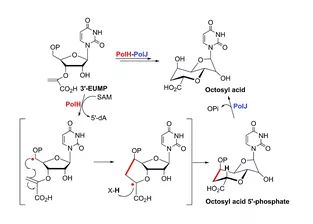
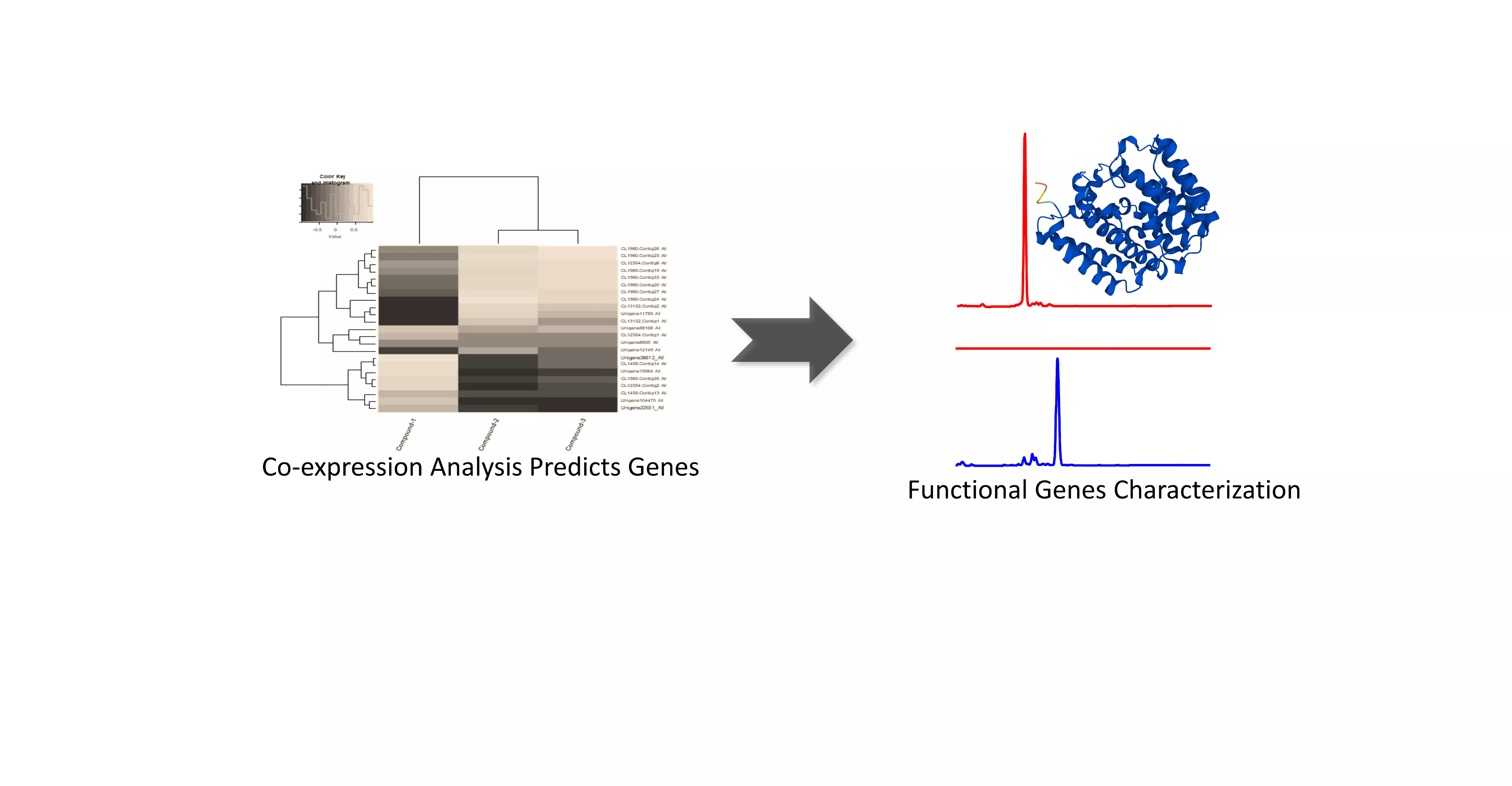
Natural product chemists have discovered numerous bioactive compounds despite the ever-decreasing discovering rates. However, only a small number of compounds are successfully developed into medicines because of the limited source of living materials, particularly marine organisms, e.g., corals, and the extremely low content of bioactive compounds inside them. Biosynthesis is one of the ways to solve this problem. And biosynthetic pathway elucidation and enzyme mechanism investigation are the foundation for achieving sustainable and scalable bioproduction (below). This leads to our second scientific mission: Understanding the mystery of natural chemical reactions by enzymes involved in the biosynthesis of bioactive compounds.
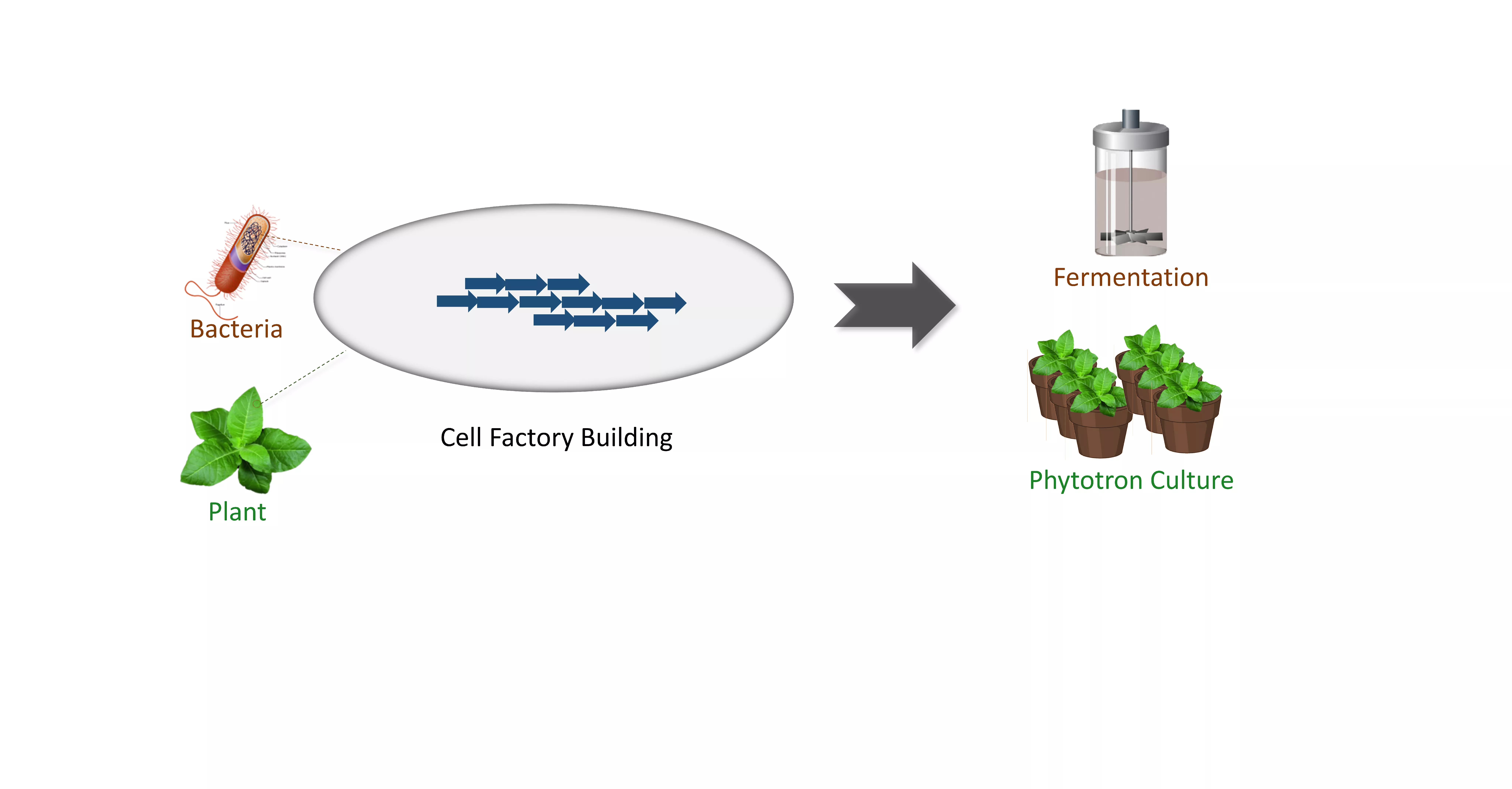
3. PRODUCE: Cell factory building for bioproduction of value-added compounds
Based on the knowledge above, we sought to construct cell factories containing a series of gene circuits using model hosts, e.g., E. coli, and tobacco, for sustainable overproduction of value-added compounds, including medicinal ones. Our lab has been equipped with 5-40 L bioreactors and a set of phytotron (~40 ㎡), facilitating the downstream overproduction-related experiments.
Students
Jin Feng Li, joint Ph.D student
Ran Zou, joint M.S. student
Xiaochen Chen, joint M.S. student
Liang Ma, M.S. student
Pantao Chu, joint M.S. student
Ting He, joint M.S. student
Jinting Wang, M.S. student
Jiaxing Chen, joint M.S. student